Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive disorder that causes muscular atrophy and hypotonia [1, 2]. In over 95% of cases, SMA is caused by homozygous deletion of exon 7 in the SMN1 gene [3-5]. However, mutations in other genes in the SMA region can contribute to the disease, such as SMN2, NAIP, and GTF2H2 genes [6-8].
The NAIP gene (OMIM: 600355) [9] is located near the SMN gene and is also duplicated in the 5ql3 region. However, the copy associated with deletions in SMA patients can be distinguished because only this copy contains exon 5 [10-12]. Therefore, in some studies, deletion of exon 5 is present in approximately 50% of patients with SMA type I and ~20% in patients with type II and type III [13]. Experiments demonstrating that the NAIP gene is responsible for expressing a protein that suppresses cellular apoptosis support the idea that the protein acts as a negative regulator of motor neuron apoptosis [14]. When this protein is deficient or absent, it contributes to the SMA phenotype [15-17]. Thus, a moderate correlation has been demonstrated between mutations in the NAIP gene and the pathophysiology of SMA, especially when considered together with the number of SMN2 copies, particularly in SMA types I, II, and III [18].
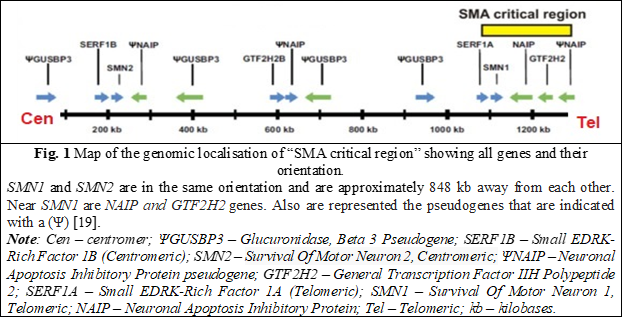
The gene GTF2H2 (OMIM: 601748) a subunit of the basal transcription factor TFIIH, involved in the transcription process, DNA repair mechanisms and probably in other cellular processes was also characterized and located in the SMA region.
Thus, studies were reported on unrelated patients with SMA that showed that large deletions involving SMN, NAIP, GTF2H2 gene loci are associated with the most severe SMA phenotype (SMA type I) [20, 21]. The identification of deletions in these genes can be performed by different molecular genetics methods such as PCR, qPCR or MLPA [13, 17, 21].
Objective
The aim of this study was to analyze the profile of modifier genes associated with SMA, such as NAIP and GTF2H2, in patients with SMA.
Material and methods
The study was conducted at the Institute of Mother and Child, Human Molecular Genetics Laboratory. A total of 105 patients suspected of having SMA were enrolled in the study, including 50 patients with a molecular-genetic diagnosis of SMA, 55 patients with hypotonia but without confirmed molecular-genetic SMA, and 107 unrelated healthy individuals from the Human Molecular Genetics Laboratory's database. All patients have signed the consent for participation (approved by Research Ethics Committee of Nicolae Testemițanu State University of Medicine and Pharmacy, Act No. 3, from February 16, 2021). Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) and multiplex ligation-dependent probe amplification (MLPA) (The innovation Act “The method of diagnosing Spinal Muscular Atrophy by the MLPA technique” No. 483 from 17.03.2022) were used to detect deletions of exons 7 and 8 of the SMN1 gene for diagnostic purposes. Multiplex polymerase chain reaction (mPCR) was used for exon 5 of the NAIP gene and exon 4 of the GTF2H2 gene. Primers for identifying mutations in exon 5 of NAIP (The innovation Act „ Diagnostic method for the deletion of exon 5 of the NAIP gene based on the PCR genetic molecular technique” No.517 from 14.03.2023) and exon 4 of GTF2H2 through mPCR (The innovation Act "Diagnostic method of deletion of exon 4 of the GTF2H2 gene based on the PCR genetic molecular technique" No. 516 from 14.03.2023) were custom-designed, along with the parameterization of the amplification program. The innovative diagnostic methods used, as outlined in the acts mentioned above, contain the detailed protocol information, safeguarding that more specific data will not be made public to facilitate the filing of a patent application. Statistical analyzes were calculated via Social Science Statistics (Online) [22].
Results
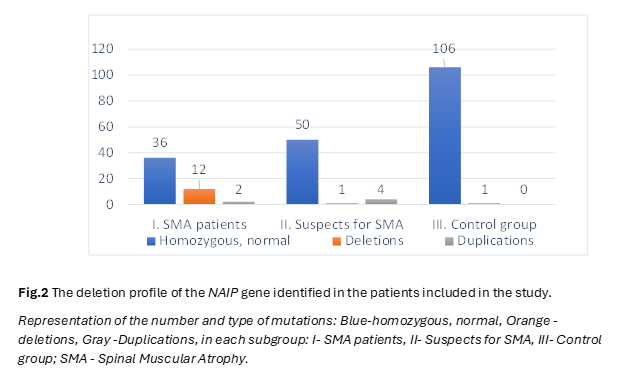
In the pursuit of unraveling the intricate genetic landscape underlying Spinal Muscular Atrophy (SMA), our study meticulously examined the distribution and implications of deletions within the SMN1, NAIP and GTF2H2 genes, elucidating potential links to SMA. The following results present a comprehensive analysis, detailing the frequency of deletions in the NAIP, GTF2H2 and SMN1 genes across distinct patient cohorts, providing numerical insights crucial for advancing our understanding of SMA pathogenesis. The distribution of patient groups was predicated on clinical manifestations and the analysis of exon 7 status in the SMN1 gene. After carrying out molecular genetics analyzes by PCR and MLPA technique, within the SMA group, 8 out of 50 patients (16%) exhibited a homozygous deletion of exon 5 of the NAIP gene, while 4 patients (8%) showed a heterozygous status, and 2 patients (4%) displayed duplications. In the cohort of 55 patients with hypotonia suspected of SMA but lacking deletions in SMN1 exon 7, one patient (2%) had a homozygous deletion of exon 5 of the NAIP gene, 3 patients (5%) exhibited a duplication of exon 5 of the NAIP gene, and one patient showcased 5 copies of the NAIP gene. Among the 107 unrelated healthy controls, one patient (1%) manifested a deletion of exon 5 of the NAIP gene (Figure 2).
Notably, the frequency of exon 5 deletions of the NAIP gene was calculated for the entire SMA patient group without categorization by SMA types.
Furthermore, molecular genetic analysis of exon 4 of the GTF2H2 gene was exclusively performed for patients with combined deletions of SMN1 and NAIP, revealing no deletions of exon 4 of the GTF2H2 gene in any patient.
Statistical analysis, employing the Chi-squared test to explore the relationship between mutations in the SMN1 gene and those in the NAIP gene, yielded compelling results. The Chi-squared statistic was X2=24.97, with an associated p-value of < 0.00001, indicating a statistically significant association between deletions in the SMN1 gene and deletions in the NAIP gene.
These findings, backed by numerical data and percentages, strongly support the hypothesis that genetic alterations in the NAIP gene are intricately linked with susceptibility to SMA. This underscores the paramount importance of delving into these genetic patterns for a nuanced understanding of SMA pathophysiology and a more accurate prognosis.
Discussions
SMA is an inherited disease that leads to progressive hypotonia and damage to the lower motor neurons in the ventral horn of the spinal cord [23]. According to previous studies, changes in genes located near the SMN gene locus have been correlated with the severity of SMA (Jiang et al., 2019) [24], and an important factor influencing the clinical severity of SMA is the duplication of exon 7/8 in the SMN2 gene, which exerts a modifying effect on the disease. Additionally, prior research in various populations has linked severity to alterations in the NAIP and GTF2H2 genes (Liu et al., 2016) [21]. Additionally, it has been demonstrated that NAIP plays a role in preventing motor neuron apoptosis and is homozygously deleted in approximately 50% of SMA type 1 cases [25], while the GTF2H2 gene is important in transcription and DNA repair [8].
Other studies, regarding the gene expressions of GTF2H2, NAIP, and others related to SMA, have been reported in the literature. For example, a study conducted in the Turkish population examined the expression levels of SERF1A, GTF2H2, NCALD, ZPR1, TIA1, PFN2, and CORO1C genes for the first time in SMA patients (Zhuri et al., 2022) showed statistically significant differences ( p = 0.037, p = 0.001) between SERF1A and NAIP genes compared between control group and patients groups [26]. Another study conducted between 2018 and 2021 included 58 SMA patients and 40 healthy individuals as a control group also in Turkish population (Karasu et al., 2022) showed that the genes NAIP (p = 0.0095) and GTF2H2 (p = 0.0049) exhibited a significant difference between healthy subjects and those with SMA [8].
In a study involving the Egyptian population, Hassan et al. in 2020 determined that SMN2 and NAIP are the primary modifier genes, and alterations in their copy numbers can impact the severity of SMA. They found that homozygous deletion of exon 5 of NAIP was observed in 60% to 73% of SMA cases, depending on the SMA type [2].
In this study, our aim was to present the mutational profile of the NAIP and GTF2H2 genes and examine the relationship between the SMN1, NAIP, and GTF2H2 genes to determine the frequency of NAIP and GTF2H2 deletions in patients with SMA. According to our findings, combined homozygous deletion in both the SMN1 and NAIP genes was found in 16% of SMA patients. Therefore, a significant association between deletions in the SMN1 gene and deletions in the NAIP gene was established. The chi-square statistic was X2 =24.97, and the p-value was <0.00001. These data once again underscore the presence of deletions in other genes in the immediate vicinity of the SMA-causing genes in the case of patients from the Republic of Moldova.
However, in our study, no relationship was demonstrated between the GTF2H2 gene and patients with deletions in SMN1 and NAIP. This phenomenon was described in a study by Arkblad et al. in 2009 [27], although the presence of mutations in the GTF2H2 gene has been reported to be closely associated with a severe form of SMA (type I) by He et al. in 2013 [28].
Due to the fact that the frequency of deletions in genes associated with SMA was calculated for the entire group of SMA patients, without categorizing them by SMA types, the percentage of deletions is different compared to other populations. However, this still demonstrates that such genetic profiles are characteristic of this disease in the Moldovan population, especially given that the p-value (<0.00001) and the chi-square test statistic (X2=24.97) showed a highly significant correlation between these two gene mutations. Regarding patients with duplication of exon 5 of NAIP, both in the SMA patient group and the hypotonia group, Tomoko Akutsu et al. in 2002 reported in their work that approximately 2 to 5 copies of intact or truncated NAIP gene have been identified in the general population, suggesting that duplications in the NAIP gene do not have clinical significance [29].
These findings suggest a complex link between SMN1, NAIP and GTF2H2 genes in the pathogenesis of SMA and highlight the importance of molecular genetic studies for understanding and characterizing the disease in different populations. Our study contributes to the knowledge of the mutational profile of the NAIP and GTF2H2 genes in the context of SMA in Moldova, while underlining the need for continued research to develop more effective therapies and to increase the understanding of this complex condition.
Some aspects that are not yet known or require investigation can be categorized as limitations but also ideas for further research:
- Perspectives on Molecular Mechanisms: Further investigations are warranted to elucidate the molecular pathways involved in establishing causality and risk factors.
- Genetic Variability: Examination of a broader spectrum of possible genetic factors implicated is essential.
- Clinical Implications: Subsequent research could shed light on how this genetic information can be practically applied in a clinical framework, with long-term monitoring of individuals with associated mutations.
Conclusions
The present study revealed a higher prevalence of NAIP gene deletions within the SMA patient group as compared to the control group, establishing a significant relationship with a p-value of p < 0.00001. This suggests that the likelihood of this relationship occurring by chance is exceedingly low. Consequently, these alterations merit consideration in the assessment of molecular pathophysiology and disease prognosis. This observation is particularly pertinent in the context of genetic therapies, as it signifies that the genetic profile characterized by modifications in genes within the SMA region is also representative of the population in the Republic of Moldova.
Competing interests
None declared.
Authors’ contribution
IC conceived conceptualization, methodology, data collection, analysis and interpretation, writing - original draft preparation. VS conceived writing review and editing, supervision, funding acquisition, validation. The authors read and approved the final version of the manuscript.
Patient consent
Obtained.
Ethics approval
This study was approved by the Research Ethics Committee of Nicolae Testemițanu State University of Medicine and Pharmacy (Act No. 3, from February 16, 2021).
Acknowledgments and funding
The present research was financed by the research project "Genomic and Metabolomic Medicine in the service of the prevention of genetic diseases for healthy generations in the Republic of Moldova" [Acronym: SCREENGEN, Cipher: 20.800009.8007.22].
Authors’ ORCID IDs
Iulia Coliban – https://orcid.org/0000-0002-8733-397X
Victoria Sacară – https://orcid.org/0000-0001-9200-0494
References
Costa-Roger M, Blasco-Pérez L, Cuscó I, Tizzano EF. The importance of digging into the genetics of SMN genes in the therapeutic scenario of spinal muscular atrophy. Int J Mol Sci. 2021 Aug 21;22(16):9029. doi: 10.3390/ijms22169029.
Hassan HA, Zaki MS, Issa MY, El-Bagoury NM, Essawi ML. Genetic pattern of SMN1, SMN2, and NAIP genes in prognosis of SMA patients. Egypt J Med Hum Genet. 2020;21(1):0-6. doi: 10.1186/s43042-019-0044-z.
de Souza Godinho FM, Bock H, Gheno TC, Saraiva-Pereira ML. Molecular analysis of spinal muscular atrophy: a genotyping protocol based on TaqMan real-time PCR. Genet Mol Biol. 2012;35(4 Suppl.):955-959. doi: 10.1590/S1415-47572012000600010.
Coliban I, Ușurelu N, Sacară V. Perspective of neonatal screening of spinal muscular atrophy. Bul Perinatol (Chisinau). 2022;1(93):166-170.
Sacară V. Particularitățile molecular-genetice ale patologiilor neuromusculare frecvent întâlnite în Republica Moldova [The molecular-genetic features of the neuro-muscular pathologies frequently encountered in the Republic of Moldova] [dissertation summary]. Chisinau: Dimitrie Cantemir State University; 2019. 42 p. Romanian.
Roy N, Mahadevan MS, McLean M, et al. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80(1):167-178. doi: 10.1016/0092-8674(95)90461-1.
Sacară V, Coliban I, Revenco N, Țurcan D, Egorov V, Ușurelu N. SMA: news and perspectives. Bul Perinatol (Chisinau). 2020;1(86):38-42.
Karasu N, Acer H, Hilal A, et al. Molecular analysis of SMN2, NAIP and GTF2H2 gene deletions and relation with clinical subtypes of spinal muscular atrophy. Res Sq. 2022:1-18. doi: 10.21203/rs.3.rs-1442537/v1.
OMIM: an Online Catalog of Human Genes and Genetic Disorders [Internet]. Baltimore: McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; 1966-2023 [cited 2023 Jul 18]. Available from: https://www.omim.org/.
Butoianu N. Atrofia musculară spinală - genetică moleculară şi mecanisme patogenice [Spinal muscular atrophy - molecular genetics and pathogenic mechanisms]. Rom J Child Adolesc Neurol Psych. 2011;(3):1-16. Romanian.
Rodrigues NR, Owen N, Talbot K, et al. Gene deletions in spinal muscular atrophy. J Med Genet. 1996;33(2):93-96. doi: 10.1136/jmg.33.2.93.
Medrano S, Monges S, Gravina LP, et al. Genotype-phenotype correlation of SMN locus genes in spinal muscular atrophy children from Argentina. Eur J Paediatr Neurol. 2016;20(6):910-917. doi: 10.1016/j.ejpn.2016.07.017.
Wadman RI, Jansen MD, Stam M, et al. Intragenic and structural variation in the SMN locus and clinical variability in spinal muscular atrophy. Brain Commun. 2020;2(2):1-13. doi: 10.1093/braincomms/fcaa075.
Burlet P, Bürglen L, Clermont O, et al. Large scale deletions of the 5q13 region are specific to Werdnig-Hoffmann disease. J Med Genet. 1996;33(4):281-283. doi: 10.1136/jmg.33.4.281.
Biros I, Forrest S. Spinal muscular atrophy: untangling the knot? J Med Genet. 1999;36(1):1-8. doi: 10.1136/jmg.36.1.1.
Jedrzejowska M, Milewski M, Zimowski J, et al. Phenotype modifiers of spinal muscular atrophy: the number of SMN2 gene copies, deletion in the NAIP gene and probably gender influence the course of the disease. Acta Biochim Pol. 2009;56(1):103-108.
Zhang Y, He J, Zhang Y, et al. The analysis of the association between the copy numbers of survival motor neuron gene 2 and neuronal apoptosis inhibitory protein genes and the clinical phenotypes in 40 patients with spinal muscular atrophy: observational study. Medicine (Baltimore). 2020;99(3):e18809. doi: 10.1097/MD.0000000000018809.
Watihayati MS, Fatemeh H, Marini M, et al. Combination of SMN2 copy number and NAIP deletion predicts disease severity in spinal muscular atrophy. Brain Dev. 2009;31(1):42-5. doi: 10.1016/j.braindev.2008.08.012.
Blatnik AJ 3rd, McGovern VL, Burghes AHM. What genetics has told us and how it can inform future experiments for spinal muscular atrophy, a perspective. Int J Mol Sci. 2021;22(16):8494. doi: 10.3390/ijms22168494.
Zeng G, Zheng H, Cheng J, et al. Analysis and carrier screening for copy numbers of SMN and NAIP genes in children with spinal muscular atrophy. Chinese J Med Genet. 2014;31(2):152-155. doi: 10.3760/cma.j.issn.1003-9406.2014.02.006.
Liu Z, Zhang P, He X, et al. New multiplex real-time PCR approach to detect gene mutations for spinal muscular atrophy. BMC Neurol. 2016;16(1):141. doi: 10.1186/s12883-016-0651-y.
Chi-Square Calculator. Social Science Statistics [Internet]. ©2023 Jeremy Stangroom [cited 2023 Aug 12]. Available from: https://www.socscistatistics.com/tests/chisquare/default2.aspx.
Shababi M, Lorson CL, Rudnik-Schöneborn SS. Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J Anat. 2014;224(1):15-28. doi: 10.1111/joa.12083.
Jiang J, Huang J, Gu J, et al. Genomic analysis of a spinal muscular atrophy (SMA) discordant family identifies a novel mutation in TLL2, an activator of growth differentiation factor 8 (myostatin): a case report. BMC Med Genet. 2019;20(1):204. doi: 10.1186/s12881-019-0935-3.
Rekik I, Boukhris A, Ketata S, et al. Deletion analysis of SMN and NAIP genes in Tunisian patients with spinal muscular atrophy. Ann Indian Acad Neurol. 2013;16(1):57-61. doi: 10.4103/0972-2327.107704.
Zhuri D, Gurkan H, Eker D, et al. Investigation on the effects of modifying genes on the spinal muscular atrophy phenotype. Glob Med Genet. 2022 Sep 5;9(3):226-236. doi: 10.1055/s-0042-1751302.
Arkblad E, Tulinius M, Kroksmark AK, et al. A population-based study of genotypic and phenotypic variability in children with spinal muscular atrophy. Acta Paediatr. 2009;98(5):865-72. doi: 10.1111/j.1651-2227.2008.01201.x.
He J, Zhang QJ, Lin QF, et al. Molecular analysis of SMN1, SMN2, NAIP, GTF2H2, and H4F5 genes in 157 Chinese patients with spinal muscular atrophy. Gene. 2013. doi: 10.1016/j.gene.2012.12.109.
Akutsu T, Nishio H, Sumino K, et al. Molecular genetics of spinal muscular atrophy: contribution of the NAIP gene to clinical severity. Kobe J Med Sci. 2002;48(1-2):25-30.